We develop and apply coarse-grained models for proteins at different levels of resolution, from detailed atomistic models to simple lattice models. Our overall goal is to discover the principles that underlie the structure, function and evolution of proteins, as well as how some molecular processes lead to disease.
Fold-switching proteins
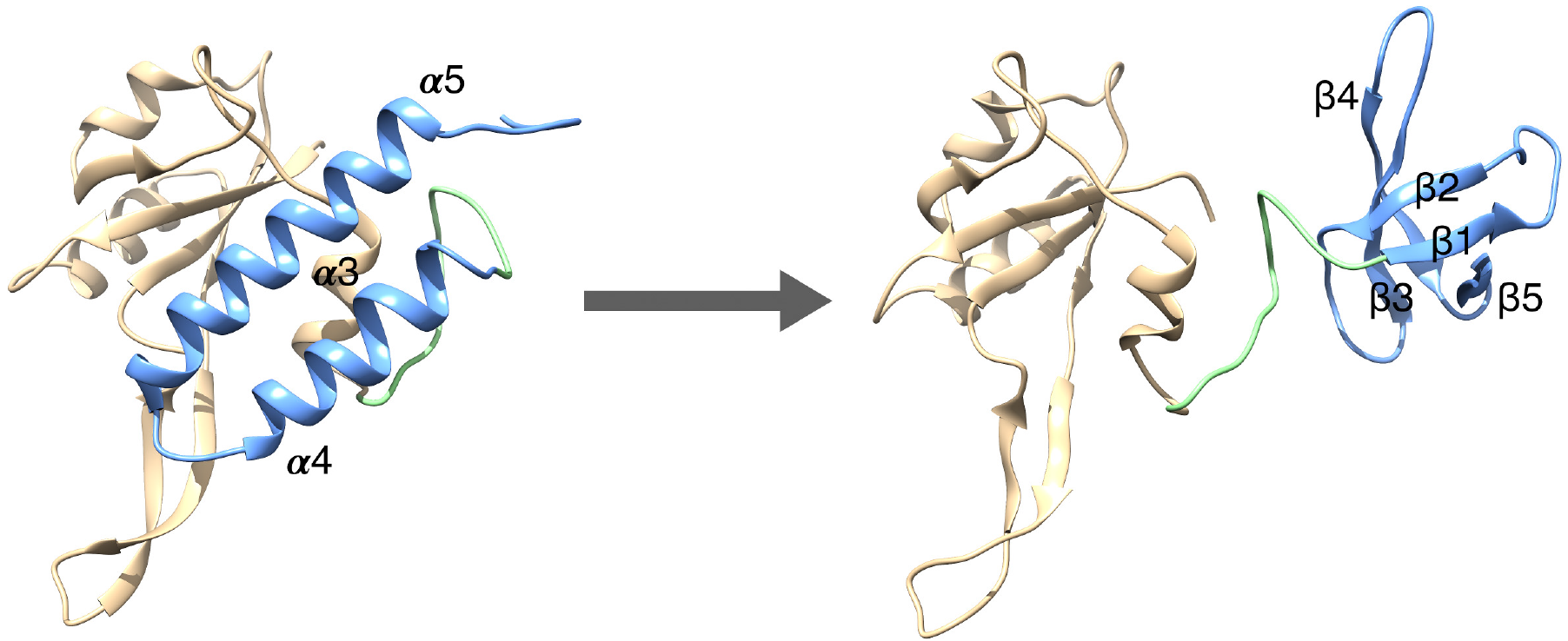 We are interested in a newly discovered class of proteins that exhibit a unique ability to reversibly switch between two entirely different folded structures. A complete understanding of these fold switching proteins could be used for biotechnological applications, such as single-molecule biosensors and targeted drug delivery.
We are interested in a newly discovered class of proteins that exhibit a unique ability to reversibly switch between two entirely different folded structures. A complete understanding of these fold switching proteins could be used for biotechnological applications, such as single-molecule biosensors and targeted drug delivery. Simulating these highly specific and large-scale fold-switching events is challenging and requires new models and conformational sampling methods. We have developed a simulation strategy that is based on an all-atom model for protein folding in combination with Monte Carlo sampling techniques.
One focus of our studies is the bacterial transcription factor protein RfaH, which exhibits one of the most dramatic fold switches discovered so far. Using our simulation approach, we have probed physical properties of this protein, including structural fluctuations and mechanical properties. We are also working on understanding the mechanism that triggers the fold switch in RfaH inside bacterial cells.
Related articles:
2) Bahman Seifi, Adekunle Aina and Stefan Wallin. Structural fluctuations and mechanical stabilities of the metamorphic protein RfaH. Proteins: structure, function and bioinformatics 89 289-300 (2021)
3) Christian Holzgräfe and S. Wallin. Smooth functional transition across a mutational pathway with an abrupt protein fold switch. Biophysical Journal 107 1217-1225 (2014).
4) Robert B. Best. Bootstrapping new protein folds. Biophysical Journal 107 1040-1041 (2014). New and Notable article.
Liquid-liquid phase separation
 Intrinsically disordered proteins, i.e. proteins that do not spontaneously fold, are prevalent in many organisms and central to many cellular processes. For example, intrinsically disordered proteins drive the formation of so-called membraneless organelles, which lack a lipid-bilayer boundaries present in organelles such as nuclei or mitochondria. Instead, these spherical membraneless organelles form spontaneously through a liquid-liquid phase transition. By condensing and dissolving depending on cellular conditions, these "droplets" give spatiotemporal control of biological reactions.
Intrinsically disordered proteins, i.e. proteins that do not spontaneously fold, are prevalent in many organisms and central to many cellular processes. For example, intrinsically disordered proteins drive the formation of so-called membraneless organelles, which lack a lipid-bilayer boundaries present in organelles such as nuclei or mitochondria. Instead, these spherical membraneless organelles form spontaneously through a liquid-liquid phase transition. By condensing and dissolving depending on cellular conditions, these "droplets" give spatiotemporal control of biological reactions. We study the properties of these liquid droplets, such as stability and concentration, and how they depend on the amino acid sequence of the disordered proteins. In particular, we have within a lattice model investigated the role of "blocks" of charged amino acids along the sequence of the disordered proteins, which are known to play an important role in the phase separation process.
Related articles:
2) Stefan Wallin. Intrinsically disordered proteins: structural and functional dynamics. Research and Reports in Biology. 8 7--16 (2017).
Other ongoing projects:
- Protein folding and macromolecular crowding.
- Coarse-graining in biomolecular simulations.
- Techniques for enhanced conformational sampling.